Literature tracking
Nearly three peer-reviewed articles that use data discovered and accessed through GBIF are now published every day of the year

The GBIF Secretariat maintains an ongoing literature tracking programme, which identifies research uses and citations of biodiversity information accessed through GBIF’s global infrastructure. We also produce summaries of recent research, many of which are then compiled in an annual Science Review.
The following describes our process for discovering, reviewing and adding a newly published article to the index.
Search strings
Using search strings combining frequently cited phrases (e.g. GBIF, Global Biodiversity Information Facility, etc.), DOI prefixes (e.g. 10.15468), URLs of IPT installations, as well as names of GBIF nodes (e.g. National Biodiversity Data Centre, CONABIO, Atlas of Living Australia, etc.), the results of daily searches are automatically fed into a sheet that serves as the raw database of potential GBIF use cases.
Sources
The articles are retrieved using alerts and XML-based feeds from the following sources:
| Source | Medium |
|---|---|
| Google Scholar | |
| Scopus | |
| Wiley Online Library | |
| SpringerLink | RSS feed |
| NCBI Pubmed | |
| bioRxiv |
Parsing raw data
New search results delivered by e-mail are piped through mailparser.io with customized rules set up for each source, ensuring a uniform and consistent output. The services uses webhooks to add an entry to a Google Sheet for each new paper containing a timestamp, the source, a title, and a URL. RSS feeds are added to the same sheet using an IFTTT applet.

Cleaning the data
The GBIF Secretariat applies the following semi-automatic steps to remove duplicates and false positives:
- First duplicate check (by title) - has the entry already been processed earlier?
- Get unique identifier (e.g. DOI) from URL or via CrossRef API
- Second duplicate check (by unique identifier) - has the entry already been added to the literature database?
- False positive check - does paper mention or cite GBIF or GBIF-mediated data?
These initial steps usually removes about half the raw input, as about 30 per cent are duplicates and 20 per cent are false positives.
Curating the data
Following clean-up, the papers are classified and categorized according to a number of criteria. Additional metadata is retrieved (primarily via CrossRef API) and stored:
- title - author names, OrcIDs and countries/territories/islands of affiliation - year of publication - journal - subject of research - source of funding - countries/territories/islands covered in research - is paper peer reviewed? - is paper open access? - number of records used/downloaded from GBIF - means of citation - download DOI, if availableStoring and indexing the data
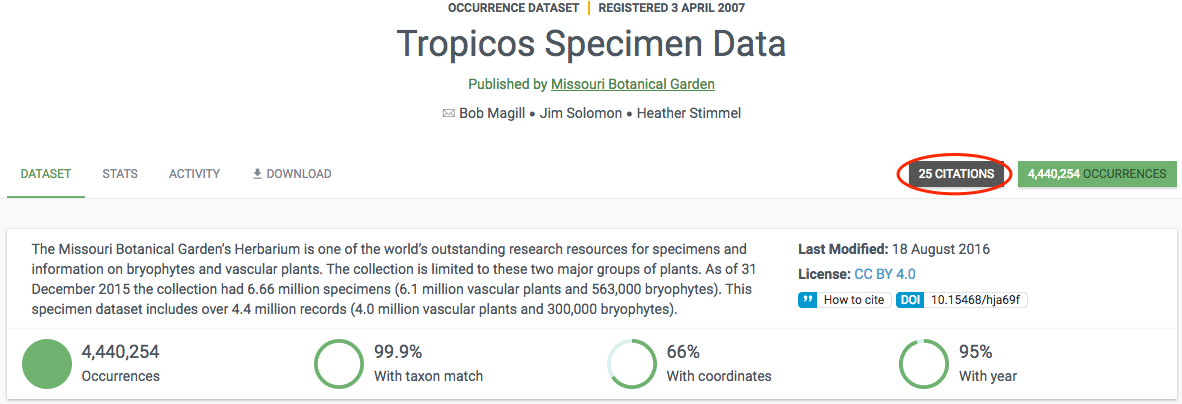
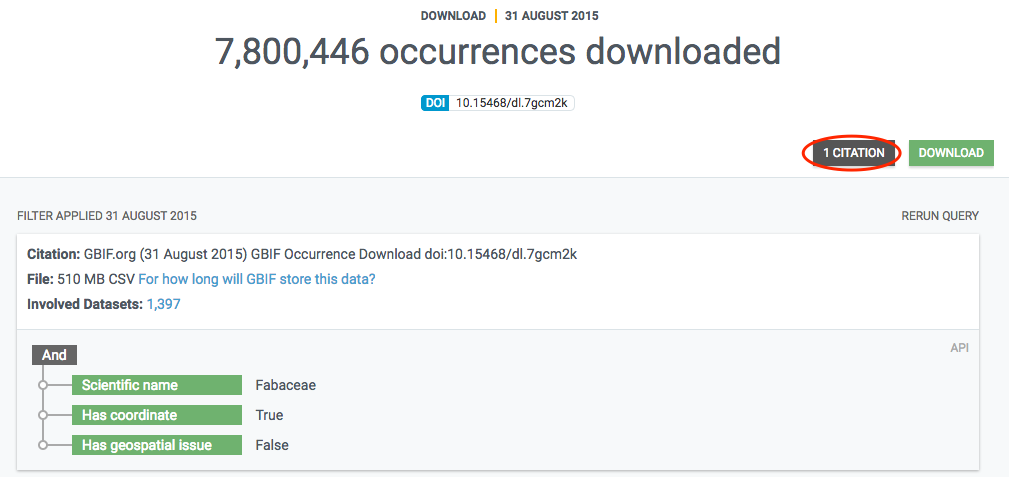
The final data is stored in a literature database (currently Mendeley) that is regularly indexed by GBIF.org and shown in the literature resource section. If a paper includes a DOI representing the GBIF-mediated data used in the study, this linkage is directly included in the index, so that datasets and publishers show links (if any) to papers that used their data. Links between research papers and data in GBIF is displayed in a number of ways on GBIF.org:
In literature searches
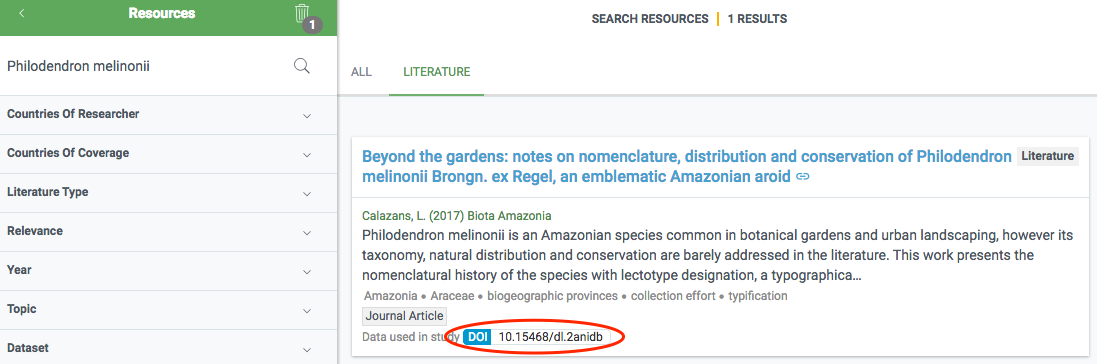
On dataset, publisher and download pages